|
Serial Block-Face SEM
by Hall, Hartwieg and Nguyen
doi:10.3908/wormatlas.9.15
Description
Scanning electron microscopy (SEM) has also undergone improvement in specimen resolution due to computer control of the specimen, better digital image
capture, better en bloc staining, and alternate methods of low voltage imaging. Thus, a well-stained sample can be viewed in scanning mode via either back-scattered electrons or secondary emitted electrons to probe within shallow distances below the specimen surface. When combined with serial thin sectioning or ion milling of the specimen surface, these imaging modalities permit Serial Block-Face SEM (SBFSEM), where block face views are collected in series in automated fashion to build a serial movie of the whole specimen by sequential erosion and imaging, step by step (Denk and Horstmann, 2004; Heyman et al., 2009; Knott et al., 2008, 2011; Mikula et al., 2012). We give a separate overview of the focused ion beam milling method for serial block-face imaging (FIB/SEM).
The SBFSEM or Denkotome consists of an ultramicrotome built inside the vacuum chamber of an SEM (Denk and Horstmann, 2004). Serial sections are removed from the block face, and the scanning head of the SEM is used to collect an image of the newly cut block face using backscattered electrons. In this manner, each image stays in exact registration, so that a serial section movie can be created through the specimen, without ever storing or imaging the thin sections themselves. While this method cannot offer equally high resolution in the Z-axis as electron tomography, it does permit extraordinary details to be acquired without mastering the manual collection and staining of serial sections. The 3D movie can then be analyzed using software packages similar to those for an electron tomogram. A commercial version of the Denkotome is available from Gatan, called the 3View. EMSerialblockFIG 1 shows sample output from a movie produced on the 3View showing the adult head of C. elegans. For a movie of the C. elegans mid-body click here or go to the WormAtlas movie gallery.
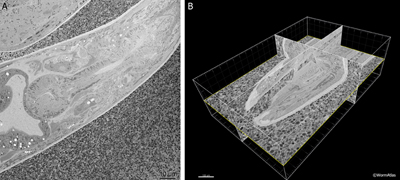 EMSerialblockFIG 1 EMSerialblockFIG 1
One oddity of the SEM method is that the block-face image collected via either secondary electrons or backscattered electrons is derived from a depth of view into the block face that is variable. Potentially one could view deeper than a typical thin section (ranging from 20200 nm) depending mostly on the accelerating voltage. Thus, each serial image could contain information regarding a rather deep portion along the Z-axis. In the Gatan 3View instrument, images are collected from backscattered electrons. At 2 keV accelerating voltage, the image is derived from about 50 nm of material closest to the block face. At 1 keV, the image may derive from only the outer 25 nm. Section thickness on the 3Views internal microtome can be set between 5 and 200 nm, but resin-embedded biological material will not cut well at less than 30 nm. The block advances via piezoelectric control, and the sections are cut on a diamond knife inside the microscope chamber under an intermediate, environmental vacuum condition. Depending on image size being collected by the SEM, the machine can cut hundreds of sections per day or more, and the 3View machine can run unattended for several weeks. One series produced in 2010 covered 6000 serial images at 2k x 2k image size, and took 2 weeks to produce under continuous operation (Joel Mancuso, Gatan; pers. communication).
Helpful Hints
At present the Gatan 3View is not equipped to do electron tomography, but we anticipate that future SEM models (from Zeiss, FEI or Gatan) will allow for the collection of several tilt images of the block face. From such tilt images, a crude electron tomogram of the block-face may add additional resolution in the Z-axis after each thin section, improving the overall resolution (Veeraraghavan et al., 2010). In the future, we anticipate continued improvements in the resolving power of the SBFSEM method, based on improvements in staining, and SEM operations.
Figures
 Click pictures for new window with figure and legend, click again for high resolution image Click pictures for new window with figure and legend, click again for high resolution image
EMSerialblockFIG 1: Three-dimensional imaging from a Denkotome. A. Sample lengthwise view of an adult hermaphrodite head. HPF/FS sample. Thin sectioned inside a Gatan 3View. Image taken using back-scattered electrons to view the block face, not the thin section. Close-packed E. coli lie outside the nematode. Scale bar, 10 µm. B. The same data are viewed from multiple angles by resampling the voxels in three dimensions. Here the original sections were collected in cross-section, but the 3D model is resampled in three different orthogonal planes. Bounding box indicates the region observed in serial images. Scale bar, 10 µm. Images provided courtesy of Joel Mancuso, Gatan Instruments.
References
Denk, W. and Horstmann, H. 2004. Serial block-face scanning electron microscopy to reconstruct three-dimensional tissue nanostructure. PLoS Biology 2, e329. Article
Hall, D.H., Hartweig, E. and Nguyen, K.C.Q. 2012. Modern electron microscopy methods for C. elegans. Methods Cell Biol. 107: 93-149. Abstract
Heyman, J.A., Shi, D., Kim, S., Bliss, D., Milne, J.L. and Subramanian, S. 2009. 3D imaging of mammalian cells with ion-abrasion scanning electron microscopy. J. Struct. Biol. 166: 17. Abstract
Knott, G., Marchman, H., Wall, D. and Lich, B. 2008. Serial section scanning electron microscopy of adult brain tissue using focused ion beam milling. J. Neurosci. 28: 29592964. Article
Knott, G., Rosset, S. and Cantonil, M. 2011. Focussed ion beam milling and scanning electron microscopy of brain tissue. J. Vis. Exp.53: e2588. Article and Video
Mikula, S., Binding, J. and Denk, W. 2012. Staining and embedding the whole mouse brain for electron microscopy. Nature Method. 9: 1198-1201. Abstract
Veeraraghavan, A., Genkin, A.V., Vitaladevuni, S., Scheffer, L., Xu, S., Hess, H., Fetter, R., Cantoni, M., Knott, G. and Chklovskii, D. 2010. Increasing depth resolution of electron microscopy of neural circuits using sparse tomographic reconstruction. IEEE Conference on Computer Vision and Pattern Recognition (CVPR), pp. 17671774. Abstract
|
